Model and Reference Population
The model used for calculating the Angus SteerSELECT genetic predictions (i.e. genomic breeding values) is based on applying SNP effects to 61,105 SNPs in a standard imputed genomic profile for each of the 9 traits, for each steer. The SNP effects resulting from univariate analyses of each trait.
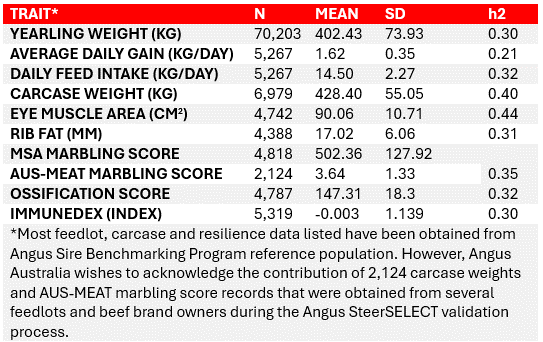
The reference population (Angus animals with genotypes and phenotypes) that underpins the Angus SteerSELECT genetic predictions is based on the comprehensive data from the Angus Australia database, including hard-to-measure traits (carcase, feed intake, immune competence) from Angus Australia’s reference population program, the Angus Sire Benchmarking program (ASBP) (Table 1). All animals in the refence population are straight-bred Angus from Australian production systems.